Welcome to ABrowse - A customizable genome browser framework!
The main features are:
- Navigate the whole genome with a Google-map-like interface.
- Create, store and share comments, annotations and bookmarks in one click.
- On-the-fly submit selected entries to Galaxy and WebLab for further analysis.
- Compatible with BioMart engine for bulk data retrieval.
- SOAP-based web service interface, fully compatible with Taverna.
- Easy to install, configure and customize for different user requirements.
- Pre-compiled binary packages and full source code are available for free download under GNU Lesser General Public License.
source codeABrowse demo instance: Arabidopsis thaliana genome browser
User Guide
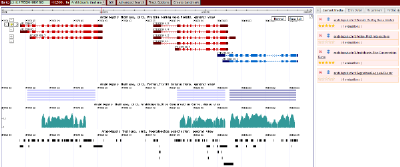
ABrowse provides different query methods for quick navigation: location search, sequence search, full text search and field specified search. User can browse query result in the center panel, smoothly draging and zooming in/out. Moreover, it's convenient to change between normal view and basepair view.
Location search
User can input an exact location to browse an interested region in two formats:
(1)chr:start-end, like "chr2:190,542-230,976".
(2)chr:start+length, like "chr2:190,542+1000".
By specifying interested annotations by setting "Track Options" switch, user can browse desired data types.
Sequence search
User can input fasta format sequences directly to initialize a search. ABrowse will map the input to the whole genome, and return the mapped locations.
- Full text search
- Full text search is supported by input concerned words directly.
- Field specified search
- Field specified search is supported for both annotation data and user-supplied comment data. User can retrieve desired data directly from this interface.
Multiple Chromosomes Annotation Browsing
ABrowse allows user to smoothly browse multiple chromosomes annotation simultaneously which is convenient for genome comparison. User can use the magic wand in the middle of navigation arrows to select a region and zoom in in the main canvas or in a sub window. Moreover, browsing tracks are listed in the "Current Tracks" panel, and user can reorder them according to comparison analysis.
Data Analysis support
- BioMart-compatible query
- User can execute BioMart-compatible batch query under the BioMart schema for data retrieval. Select a dataset and interested fields, then set the query conditions, user can get the desired result.
- Web Service query
The main aim of ABrowse Web Service is to provide programmatic access to the underlying database in order to help users in integrating ABrowse into their applications. To ensure software from various sources work well together, this integration technology is built on open standards such as Simple Object Access Protocol (SOAP), a messaging protocol for transporting information, Web Services Description Language (WSDL), a standard method of describing Web Services and their capabilities. Currently, ABrowse Web Service is compatible with Taverna workbench of myGrid project.
The ABrowse WSDL specifies the contract between external developers and the ABrowse Web Services. The Web Service Description Language (WSDL) is an XML format for describing network services. And a WSDL binding describes how the service is bound to a messaging protocol, particularly the SOAP messaging protocol. The ABrowse WSDL file is:Web Service WSDL File.
Interface Description:
Method Name Parameters Result Type
- getGenomes() No parameter String Array of all genomes
- getTracks() String genome String Array of all tracks
- getAnnotationNumberFromRegion() String genome, String track, String chromosome, String start, String end, String length_greater_than, String length_less_than, String strand Integer of result number
- getAnnotationFromRegion() String genome, String track, String chromosome, String start, String end, String length_greater_than, String length_less_than, String strand Two-dimension String Array of annotations (maximum results upto 2000 entries at a time)
- getAnnotationFromID() String genome, String track, String ID Two-dimension String Array of annotations (maximum results upto 2000 entries at a time)
- getComment() String genome, String track, String entryID, String account Two-dimension String Array of comments (maximum results upto 2000 entries at a time)
- fullTextSearch() String queryString Two-dimension String Array of annotations (maximum results upto 2000 entries at a time)
Web Service interface JavaDoc: ABrowse interface JavaDoc
Test Case (in Java code): ABrowseWSClient.tar.gz
Test case in soapUI can be found in the soapUI test page.
Computational analysis support
All the sequence data and query data in ABrowse can be sent to external bioinformatic analysis platform WebLab or Galaxy for further computation in one click. User can submit data in four ways:
(1)Select a region in the graphic chromosome and submit the selected sequence. (Click the magic wand to activate selecting status, then click a point as the start point and release the mouse. Now move to the end point and click it, then the rectangle area between the start point and end point is selected.)
(2)Submit nucleotide/protein/genomic sequence in "Entry Detail" directly.
(3)Submit BioMart batch query result directly to external bioinformatic analysis platforms.
(4)Submit comment search result directly to external bioinformatic analysis platforms.
User Registration & User Space
ABrowse provides user space for customized data.
User registration
User can register a ABrowse account freely. As a registered user, user can add own annotaion, make track evaluation, save important analysis status and save sequence query result.
Make evaluation for track
ABrowse allows user to make track evaluation by adding stars and comments. One user can add comments many times, while only one time for star evaluation.
Add comment for entry
User can add comments for entries with private/public privilege. If it's private, then only the contributor can read it. If it's public, then everybody can read it. Therefore public comments are useful for sharing and exchanging knowledge among members, while private comments can be taken as personal online work notes.
Besides pre-computed entry, ABrowse provide "My Instant Note" (Demo Video) for every registered user, supporting user to select a region on-the-fly and attach comment for it. Then user can add comment for any length of region freely, ranging from a single nucleotide SNP position to a long QTL region. (Click the magic wand to activate selecting status, then click a point as the start point and release the mouse. Now move to the end point and click it, then the rectangle area between the start point and end point is selected. Choose the last item "Add region to my instant note", and add comment to the region, then it will be added in the "My Instant Note" track instantly.)
Browsing history record
By recording browsing status as a "landmark", user can quickly jump to preserved important analysis status. Moreover, login user can share landmark with specified users. (This function is available for both registered user and guest.)
Sequence query result record
Sequence search results can be saved automatically for further analysis. (This function is available for both registered user and guest.)
Upload & Manage My Tracks
Login user can upload own data from web interface. After setting the track name, genome name, file format, privilege and file path, user can upload local file to the ABrowse genome browser instantly and do further analysis under the context of the other tracks. Uploaded tracks will be listed in the "My Tracks" panel, and user can delete it if it's unnecessary.
Tips:
ABrowse supports data transport in compression files. Please set the "network.http.accept-encoding" to "gzip,deflate" in your browser to enable compression files accepting, which will speed up your browsing.

We create the perfect
tailored solution for you
Secured
Tell what's the value for the
customer for this feature.
All in one
Write what the customer would like to know,
not what you want to show.
Easy to use
A small explanation of this great
feature, in clear words.
Useful options
Beautiful snippets
Amazing pages
Outstanding images

Our mission
Provide clear and concise financial advices to make your financial wealth grow. And this, completely free of charge.

Our values
We created our company to improve the work-life balance. We believe that we need to focus on our family more than our work and finances.

Our team
Composed of 25 people from 25 to 66 years, we’re a young and dynamic company. We have open positions, do not hesitate to contact us.